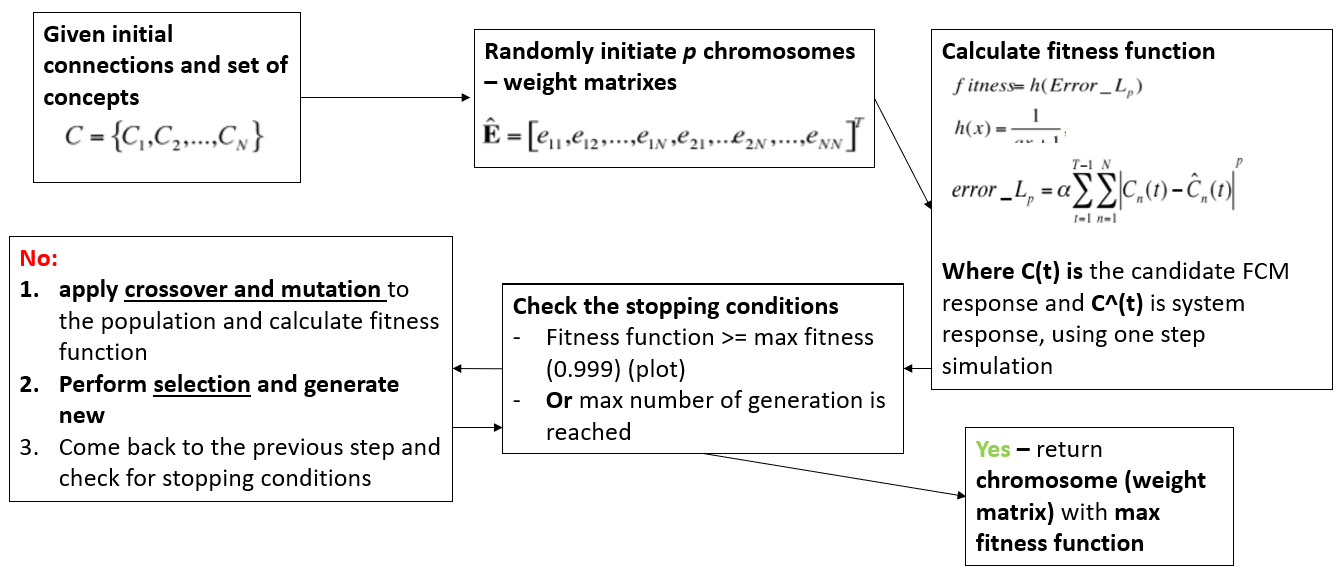
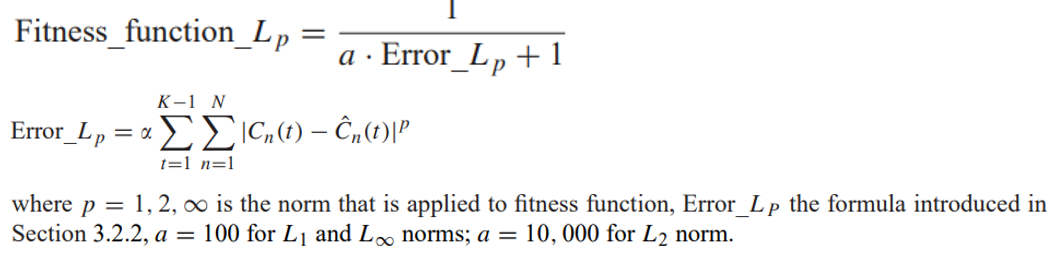RCGA¶
Task: create FCM based on one input¶
Step 1. Crossover¶
- Given two chromosomes and their weights matrixes, we randomly switch n weights between these matrixes
- Many crossover methods are available
- used with probability of 90%

Step 2. Mutations¶
Value of randomly chosen weight in the chromosome is being change into new value with the same sign
Step 3. Selection¶
Selection is the stage of a genetic algorithm in which individual genomes are chosen from a population for later breeding (using the crossover operator).
Roulette wheel selection
Selecting with weight probability, depending on the fitness of the individual gene Fitness values [1, 2, 3, 4], then the sum is (1 + 2 + 3 + 4 = 10). Therefore, you would want the probabilities or chances to be [1/10, 2/10, 3/10, 4/10] or [0.1, 0.2, 0.3, 0.4] The chosen one is taken for the crossover
Tournament selection
Winner of the tournament is being selected for the crossover two variants of the selection: with and without replacement
Step 4. Fitness function¶
Fitness is calculated as difference between concept values on each step of the simulation
For more information about the algorithms, please check out LEARNING AND AGGREGATION OF FUZZY COGNITIVE MAPS – AN EVOLUTIONARY APPROACH by Wojciech Stach